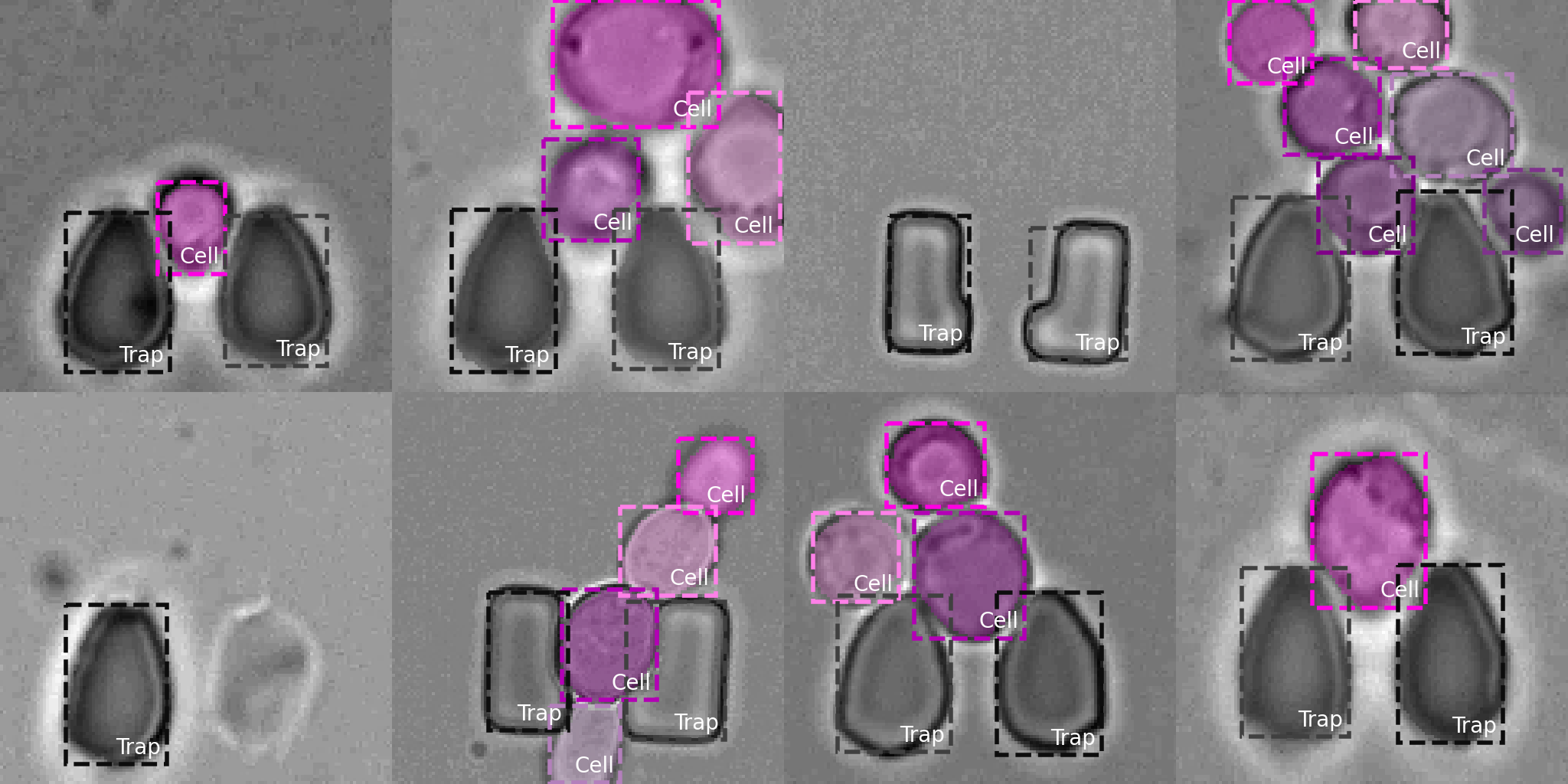
An Instance Segmentation Dataset of Yeast Cells in Microstructures
EMBC 2023
Department of Electrical Engineering and Information Technology,
Technische Universität Darmstadt
2Aeronautics Institute of Technology (work done while at TU Darmstadt)
Abstract
Extracting single-cell information from microscopy data requires accurate instance-wise segmentations. Obtaining pixel-wise segmentations from microscopy imagery remains a challenging task, especially with the added complexity of microstructured environments. This paper presents a novel dataset for segmenting yeast cells in microstructures. We offer pixel-wise instance segmentation labels for both cells and trap microstructures. In total, we release 493 densely annotated microscopy images. To facilitate a unified comparison between novel segmentation algorithms, we propose a standardized evaluation strategy for our dataset. The aim of the dataset and evaluation strategy is to facilitate the development of new cell segmentation approaches.
Dataset
We present and publicly release (under MIT license) a dataset of yeast (Saccharomyces cerevisiae) cells in microstructures with instance segmentation annotations. The dataset is comprised of 493 densely annotated brightfield microscopy images (instance segmentation) of different TLFM experiments with different microstructures (traps).Table 1. Training, validation, and test split of our yeast cells in microstructure dataset.
We also present a unified validation approach to facilitate a fair comparison for novel cell segmentation approaches. The validation code and dataset classes are available (under the MIT license) on GitHub. The dataset can be downloaded from TUdatalib and is released under the MIT license. Alternitivle you can just use wget.
wget https://tudatalib.ulb.tu-darmstadt.de/bitstream/handle/tudatalib/3799/yeast_cell_in_microstructures_dataset.zip
For details on the dataset format and examples on data loading, visualization, and validation please have a look at the GitHub repo.
Citation
If you use our dataset or find this research useful in your work, please cite the following papers:Additional Unlabeled Data
We also offer additional unlabeled microscopy images of yeast cells in microstructures which can be used for unsupervised pre-training (or similar). In the paper Multi-StyleGAN: Towards Image-Based Simulation of Time-Lapse Live-Cell (MICCAI 2021) we proposed ~9k unlabeled microscopy images (sequences). The dataset is available at TUdatalib. Please cite the following paper if you are using the unlabeled images in your research (BibTeX reference).Acknowledgements
We thank Christoph Hoog Antink for insightful discussions, Klaus-Dieter Voss for aid with the microfluidics fabrication, Jan Basrawi for contributing to data labeling, and Robert Sauerborn for aid with setting up this project page.
This work was supported by the Landesoffensive für wissenschaftliche Exzellenz as part of the LOEWE Schwerpunkt CompuGene. H.K. acknowledges the support from the European Research Council (ERC) with the consolidator grant CONSYN (nr. 773196). C.R. acknowledges the support of NEC Laboratories America, Inc.
|
Design / source code from Jon Barron's Mip-NeRF / Michaël Gharbi's website Copyright © Christoph Reich 2023 |